on the April 3, 2024

A study published by Michèle Studer's team at the Institut de Biologie Valrose in the journal Protein Science in April 2024, proposes a new approach to understanding the molecular basis of genetic diseases, by linking specific mutations in the NR2F1 gene to the varied symptoms of BBSOAS patients. These advances could also have important applications for the study of other genetic diseases.
To improve the accuracy of diagnosis and treatment, it is crucial to understand the relationship between genetic mutations and disease symptoms. In particular, this requires a thorough understanding of how mutations affect protein structure and function.
In this study, the researchers used an approach combining computer simulations of the protein structure of different genetic mutants of NR2F1 with cellular experiments to investigate the effects of mutations on cell proliferation and survival. They also employed an innovative synthetic biology technology called "genetic code expansion" (GCE) to introduce unconventional amino acids into the NR2F1 protein. By incorporating a photocrosslinking amino acid capable of forming covalent bonds with neighboring molecules when exposed to UV light, the researchers were able to visualize the protein interactions of NR2F1 and its various mutants with unprecedented precision. This approach revealed how NR2F1 interacts with other proteins, and how these interactions can be disrupted by genetic mutations. In addition, this method identified a new protein partner of NR2F1, CRABP2, which plays a role in its cellular localization in response to specific signals.
In summary, this study proposes a new approach to understanding the molecular basis of genetic diseases, by linking specific mutations in the NR2F1 gene to the varied symptoms of BBSOAS patients. These advances could also have important applications for the study of other genetic diseases.
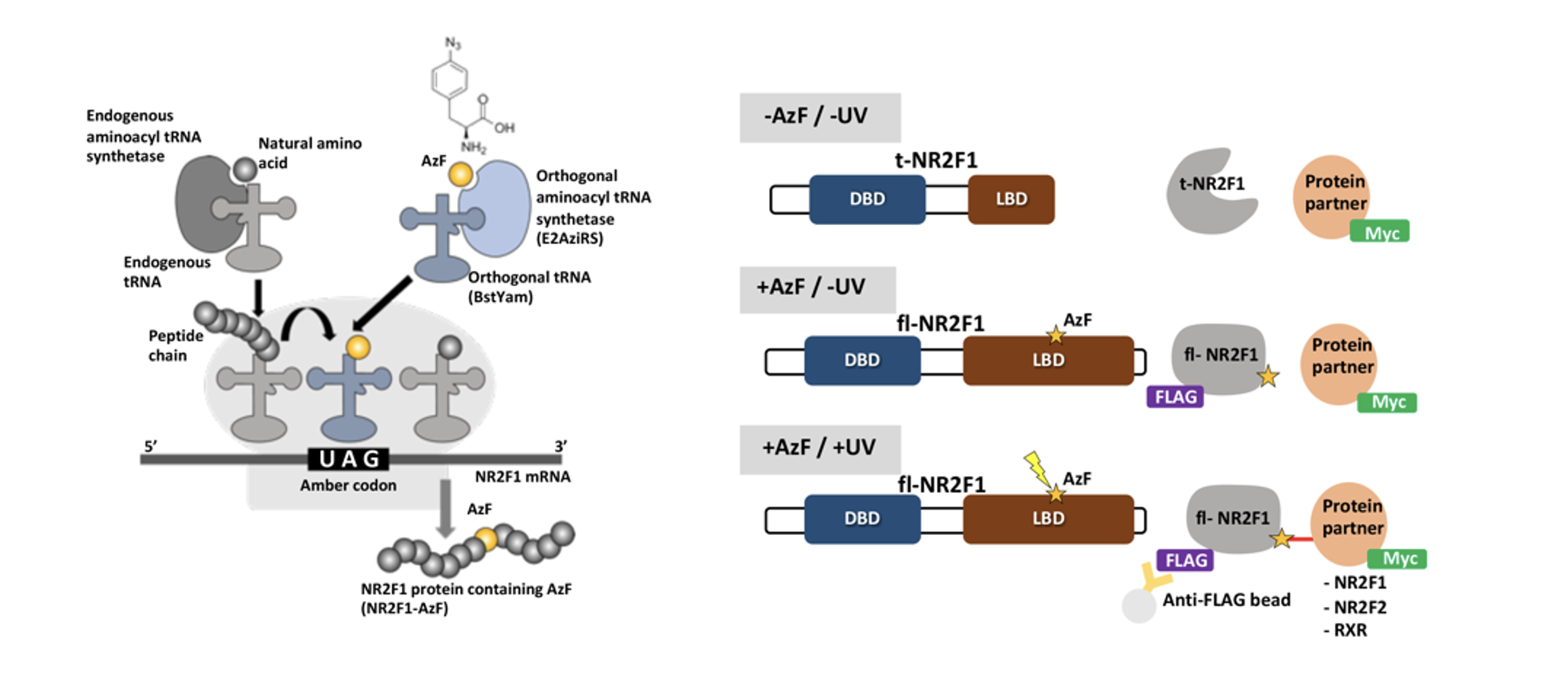
Left: Principles of site-specific incorporation of photocrosslinking amino acid by amber codon suppression. Endogenous aminoacyl tRNA synthetase charges the endogenous tRNA with its cognate amino acid. Aminoacyl-tRNA enters the ribosome and adds the amino acid to the corresponding codon. For the AzF incorporation in NR2F1, the orthogonal aminoacyl tRNA synthetase (E2AziRS) catalyzes the aminoacylation between AzF and the orthogonal tRNA (BstYam). The AzF-charged tRNA enters the ribosome and incorporates the AzF in response to the designated amber codon. The translation continues and produces full-length, AzF-containing NR2F1 protein. Right: Diagram showing UV-induced, site-specific crosslinking via AzF. NR2F1 is produced as a truncated form (t-NR2F1) in the absence of AzF due to the designated amber codon. In the presence of AzF, the amber codon suppression allows the incorporation of AzF at the selected amber codon position and the NR2F1 protein carrying AzF is translated into the full-length FLAG-tagged form (fl-NR2F1). Upon UV irradiation (365 nm), AzF forms a covalent bond between FLAG-NR2F1 and Myc-tagged putative partners (NR2F1, NR2F2, RXRa). The proteins in the crosslinked complex can be co-IP with NR2F1 by using anti-FLAG antibody-conjugated beads and detected in a higher molecular weight complex by immunoblot. See also Marino et al, Protein Sci. 2024.”
- To find out more
-
https://onlinelibrary.wiley.com/doi/10.1002/pro.4953
Marino V, Phromkrasae W, Bertacchi M, Cassini P, Chakrabandhu K, Dell'Orco D, Studer M.
"Disrupted protein interaction dynamics in a genetic neurodevelopmental disorder revealed by structural bioinformatics and genetic code expansion". Protein Sci. 2024 Apr;33(4):e4953. doi: 10.1002/pro.4953. - Author contacts :
-
Michèle Studer | INSERM Research Director +33 489150720 | Michele.studer@univ-cotedazur.fr



